|
Michigan Branch Spring 1999 Meeting
The Spring Meeting of the Michigan Branch of the American Society for Microbiology
will be held on Saturday April 10, 1999 in the McKenny Union of Eastern
Michigan University in Ypsilanti, Michigan. Five research lectures
are planned including an ASM
Foundation Lecture by Leona Ayers. The basic theme of the meeting is
"Trends in Infectious Diseases" and will provide current information for
educators, researchers, and clinicians. In addition, student research posters
are invited. The meeting is being organized by James VandenBosch. In addition
to the presentations, the annual Michigan Branch business meeting will
be held during the luncheon.
Meeting Agenda:
Please note that the presentation by the Foundation for Microbiology
Lecturer, Leona Ayers, will be held at the Halle Library Auditorium. The
Halle Library is a short walk from the McKenny Union. The lecture is in
the library so that it can be teleconferenced with the Upper Peninsula
Twig which is having their first
meeting at the same time but at Michigan Technological University in
Houghton.
| 8:15 - 9:00 |
Registration |
Continental Breakfast |
McKenny Union
Upper Level Lobby |
| 9:00 - 9:05 |
Jim VandenBosch |
Welcome |
McKenny Union Alumni Room |
| 9:05 - 9:50 |
Ray Podzorski |
HIV Genotyping: Trenches to Benches |
McKenny Union Alumni Room |
| 10:00 - 10:45 |
Carol Kauffman |
Trends in Opportunistic Fungal Infections |
McKenny Union Alumni Room |
| 11:00 - 11:45 |
Leona Ayers |
Infectious Disease Diagnosis: Are We Ready for the Third Millenium? |
Halle Library Auditorium |
| 11:45 - 12:00 |
Branch Business Meeting |
|
Halle Library Auditorium |
| 12:00 - 1:00 |
Luncheon |
|
McKenny Union
Tower & Faculty Rooms |
| 1:00 - 1:45 |
Frances Downes |
Foodborne Diseases: Recent Trends and Control Efforts |
McKenny Union Alumni Room |
| 2:00 - 2:45 |
Alan Hudson |
Chlamydia pneumoniae and Alzheimer's Disease |
McKenny Union Alumni Room |
The Speakers:
|
ASM Foundation Lecturer
Leona
Ayers
Professor of Pathology, Ohio State University
Medical Director, Ohio Department of Health Laboratories
|
Infectious Disease Diagnosis: Are We Ready for the Third Millenium? |
|
Frances
Pouch Downes
Acting Laboratory Director,
Michigan Department of Community Health
|
Foodborne Diseases: Recent Trends and Control Efforts |
|
Alan Hudson
Associate Professor, Department of Immunology and Microbiology, Wayne
State University School of Medicine, Detroit, MI
|
Chlamydia pneumoniae and Alzheimer's Disease |
|
Carol
Kauffman
Chief of Infectious Diseases, Veterans Affairs Health System, Ann Arbor,
MI
Professor, Department of Internal Medicine,
The University of Michigan School of Medicine
|
Trends in Opportunistic Fungal Infections |
|
Ray Podzorski
Assistant Professor, Department of Pathology,
Wayne State University School of Medicine, Detroit, MI
|
HIV Genotyping: Trenches to Benches |
The Posters:
Students are invited to present posters. These will be added as the
titles are received.
| The Identification of a New Family of Sugar Efflux Pumps in Escherichia
coli
J. Y. Liu, P. F. Miller*, M. Gosink, and E. R. Olsen
Parke-Davis Pharmaceutical Research, Division of Warner-Lambert Company,
Ann Arbor, MI 48106-1047
*Pfizer Central Research, Groton, CN 06340
|
Using a functional cloning strategy with an Escherichia
coli genomic plasmid library, we have identified a new family of sugar
efflux proteins with three highly homologous members in the E. coli
genome. In addition, two open reading frames, one present in Yersinia
pestis and the other in Deinococcus radiodurans, appear to encode
closely related proteins. An in vitro transport assay using inside-out
membrane vesicles prepared from overproducing strains demonstrated that
members of this new family can efflux [14C]-lactose
and [14C]-glucose. Substrate specificity
was further explored using both the inhibition of the in vitro [14C]-lactose
transport assay and a cell based assay. The results indicate that one member
of this family (SetA) prefers glycosides with alkyl or aryl substituents
as both isopropyl-ß-D-thiogalactoside (IPTG) and o-nitrophenyl-ß-D-thiogalactoside
are substrates for efflux. As sugar efflux phenomena have been reported
previously in several bacterial species including E. coli, the identification
of a new family of sugar efflux proteins may help reveal the physiological
role of sugar efflux in metabolism. It is proposed that the E. coli
members of this family, whose functions were previously unknown, be given
the gene family designation set for sugar efflux
transporter. |
| DNA Sequence Comparison of Chromosome- and Plasmid- encoded mupA
in Staphylococcus aureus.
M. Raynor*, J. E. Patterson, D. R. Reagen,C. Kauffman, and T. M. Morton.
Eastern Michigan University, Ypsilanti, MI, 48197
Veterans Affairs Health System, Ann Arbor, MI,
|
The mupirocin resistance gene, mupA, is associated with mupirocin resistance
in Staphylococcus aureus. High-level mupirocin resistance is due to expression
of a plasmid-encoded mupA. Since only high-level resistant S. aureus strains
have been associated with clinical and microbiological failure, low-level
resistant strains have not been considered clinically important. Recent
reports have identified, by Southern blot analysis, a chromosome-encoded
mupA in some low-level resistant strains of S. aureus. In order to determine
if the chromosome-encoded mupA in low-level resitant strains was identical
to the plasmid-encoded mupA in high-level resistant strains, a 1.65 kb
internal fragment was amplified by the polymerase chain reaction and DNA
sequence analysis completed. Chromosomal DNA from four different low-level
mupirocin resistant S. aureus clinical isolates, and plasmid DNA from three
high-level mupirocin resistant isolates was amplified, sequenced and compared
to the published mupA sequence. The S. aureus strains were from three different
geographic locations. The amplified DNA of all strains was virtually identical
to the published mupA sequence (>98% identity). Therefore, the chromosome-encoded
mupA in the low-level mupirocin resistant S. aureus strains studied is
identical to the plasmid encoded mupA in high-level resistant strains,
and may represent a mechanism for the dissemination of high-level mupirocin
resistant in S. aureus. |
| Immuno-chemical characterization of 5-aminolevulinic acid synthase
isoenzymes (ALAS-A and ALAS-T) of Rhodobacter sphaeroides 2.4.1.
Luiza Kryszak* and Jill Zeilstra-Ryalls
Oakland University, Rochester, MI 48309-4476
|
Antisera were raised against the two ALA (5-aminolevulinic acid) synthase
isozymes, ALAS-A and ALAS-T, of the photosynthetic bacterium, Rhodobacter
sphaeroides. The reactivity of each sera towards its cognate protein was
confirmed by parallel immunoblot analysis of Escherichia coli strains expressing
fusion polypeptides comprised of portions of ALA synthase and glutathione-S-transferase
(GST), using the ALAS antisera and anti-GST antibodies. Further analyses
demonstrated that each ALAS antiserum not only binds to both ALAS isozymes
(albeit with higher affinity towards its cognate enzyme), but also binds
to mouse ALAS. Immunoblot analysis of ALAS-A and ALAS-T overproducing strains
of E. coli revealed that the majority of each protein is produced in an
insoluble form, suggesting one explanation for the low yield of active
enzyme in the case of ALAS-A, and the inability to recover any active enzyme
in the case of ALAS-T. The antisera has already proven useful in studying
ALAS levels in R. sphaeroides, providing evidence that ALAS-T may be regulated
post-transcriptionally, a novel and important contribution to our long
term goal of understanding the regulation of expression of the two ALA
synthase genes in this organism. |
|
|
Registration Details:
Registration for the meeting can be done two ways. Registration can
be done electronically by completing the Meeting
Registration Form. Registration can also be done by completing the
form in THE SCOPE which will be mailed the first week of March. The form
in THE SCOPE should be returned to the meeting organizer, Jim VandenBosch,
at the address listed below. All registration requests received by March
27, 1999 will receive the Luncheon included in the registration fee. Requests
received after March 27 are not guaranteed a meal. Corporate sponsors should
contact Michael Huband the
Exhibitor and Meeting Program Contact.
Inquiries and Reservations:
James L. VandenBosch
Department of Biology
316 Mark Jefferson
Eastern Michigan Univaersity
Ypsilanti, MI 48197
telephone: (734) 487-4432
fax: (734) 487-9235
e-mail: [email protected]
Fees:
Please note that membership fees are $3/year for Students
and $10/year for full Members.
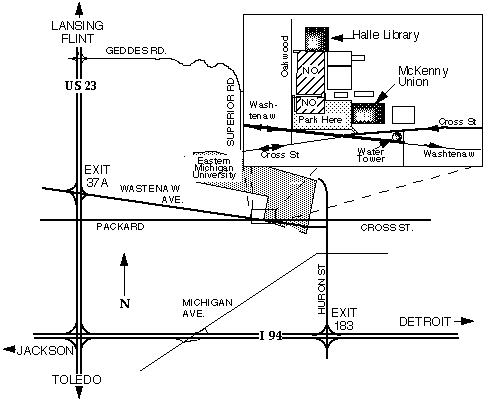
Directions:
From Detroit or Jackson: Take I94 to Ypsilanti
exit #183. Go north on Huron Street. Staying in the middle lane, continue
approximately 1 mile through three traffic lights. Move into the left lane
and it will turn left onto Cross Street just before a stop sign. Continue
on Cross Street staying in the right lane through several more traffic
lights. Immediately after the water tower (you can't miss it) turn right
into the parking lot. The meeting is in the McKenny Union, the three story
brick building adjoining the parking lot.
From Toledo, Flint & Lansing: Either take US23 to I94 east
and follow the instructions from Detroit and Jackson or take the Washtenaw
Ave. (37A) exit off of US23. If you take the Washtenaw Ave. exit (37A),
continue east on Washtenaw approximately 3 miles until you come to the
water tower (you can't miss it). Make two left turns (essentially making
a U-turn around the water tower) and turn right into the parking lot. The
meeting is in the McKenny Union, the three story brick building adjoining
the parking lot.
|



